

|
|
|
BufAMPpred: Deep Learning based AMP Prediction Server for Buffalo is the web server designed to predict if a peptide sequence has antimicrobial properties or not.
The webserver is developed based on a Convolutional Neural Network (CNN) along with
Long Short Term Memory (LSTM) based classification predictor for the classification of AMPs. |
|
The BufAMPpred contains five tabs viz. Home, Analysis, Tutorial, Team and BufAMPpred. |
|
Home -> This tab provides the brief information of BufAMPpred server. |

|
|
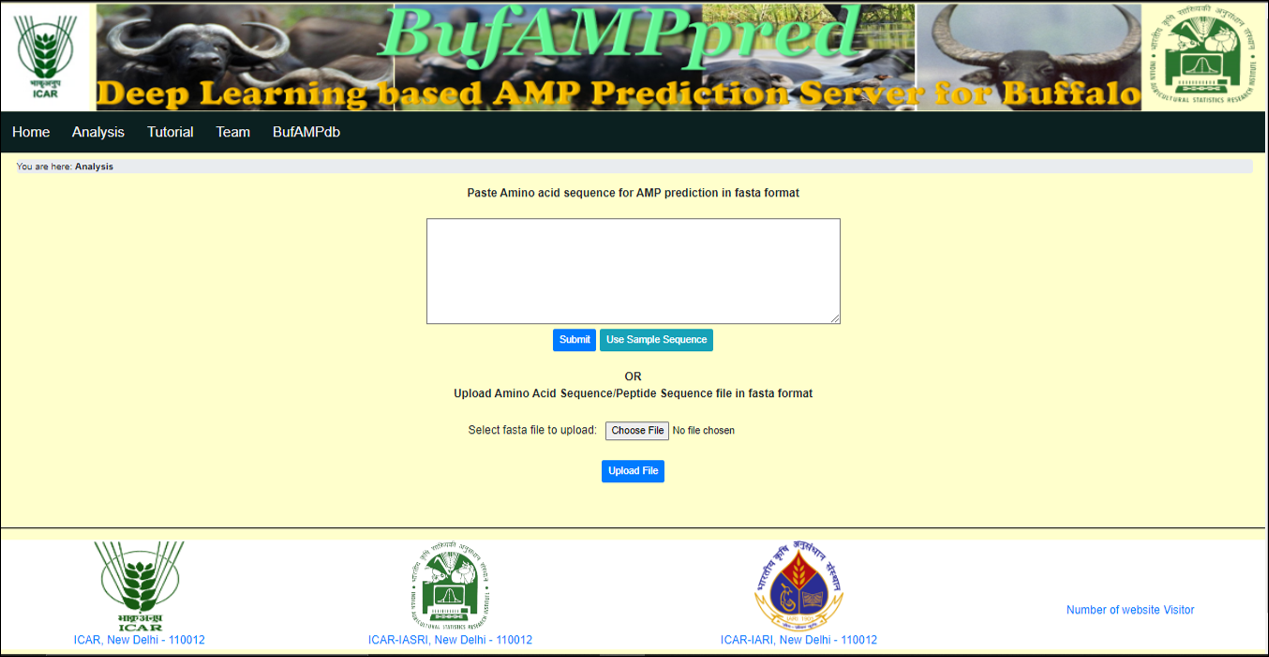
Analysis -> This tab provides the facility to analyse the uploaded fasta file (Amino acid sequence / Peptides sequence) and give the result in a tabular form. |
|
|
Input fasta file (Peptides sequence/Amino acid sequence) can directly be imported to the web server in two modes: a. Direct from the input insertion box on the web page b. Via uploading the file (fasta file) |

|
|
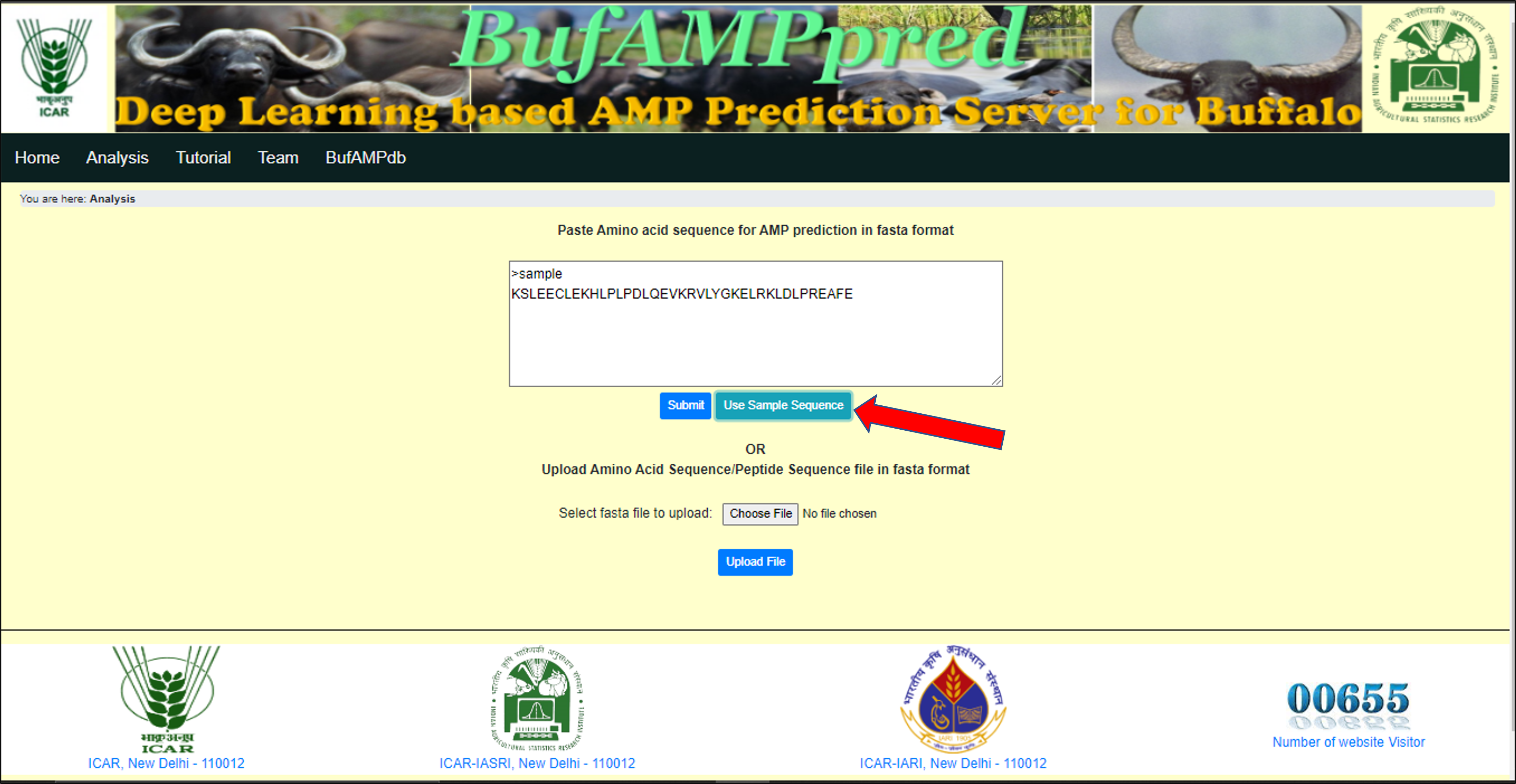
User can also see the fasta format by clicking on the Use Sample Sequence button and it will automatically fill the input insertion box on the web page. |
|
|
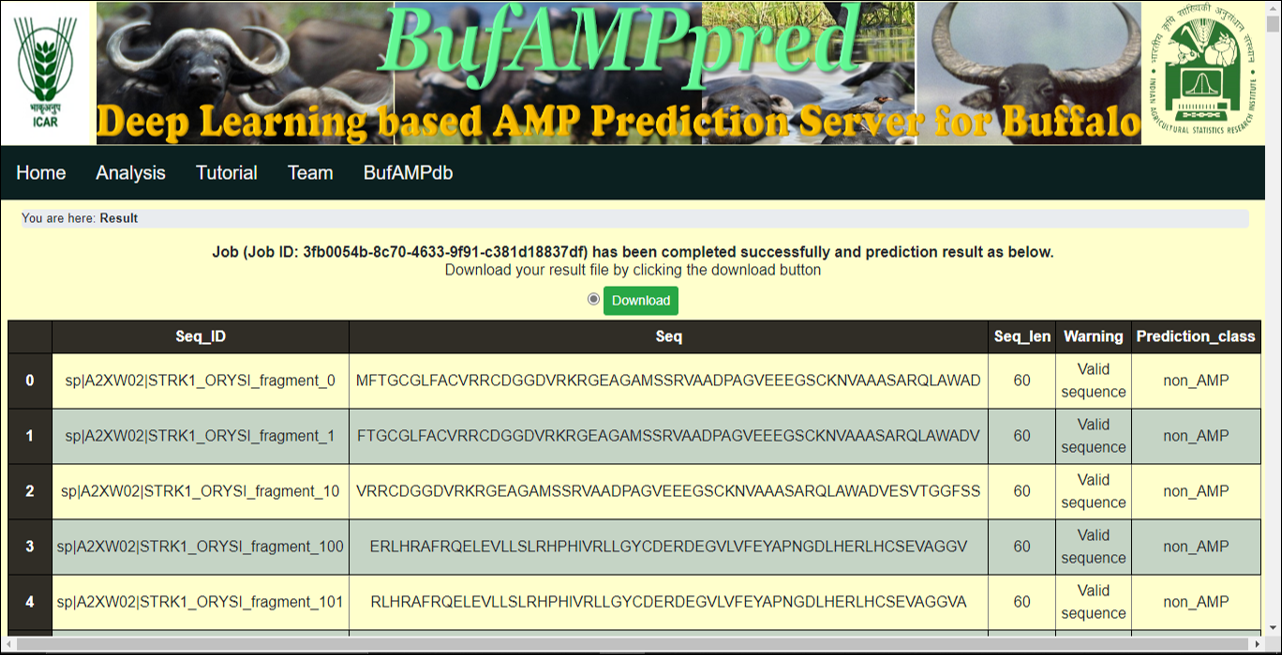
Result -> click on the Submit button after inserting fasta file in the input insertion box on the web page or click on the Upload File button after choosing fasta file from your system, it will show the result of the given fasta file in a tabular form along with prediction class of the AMP or nonAMP sequence. The Result can be easily downloaded in csv format by clicking on the Download button. |
|
|
BufAMPdb -> This tab redirects the users to the link of BufAMPdb which provides collection of all candidate AMPs (61711) and non-AMPs (2529971) predicted for buffalo along with their corresponding gene information by using the BufAMPpred. |
NOTE: The prediction of the Amino acid sequences or Peptide sequence is done only by taking the 60 length fragment of the input sequence. The sequences greater than 60 in length will be cut down in 60 length fragments by sliding one length at a time and will make fragments of the large file.